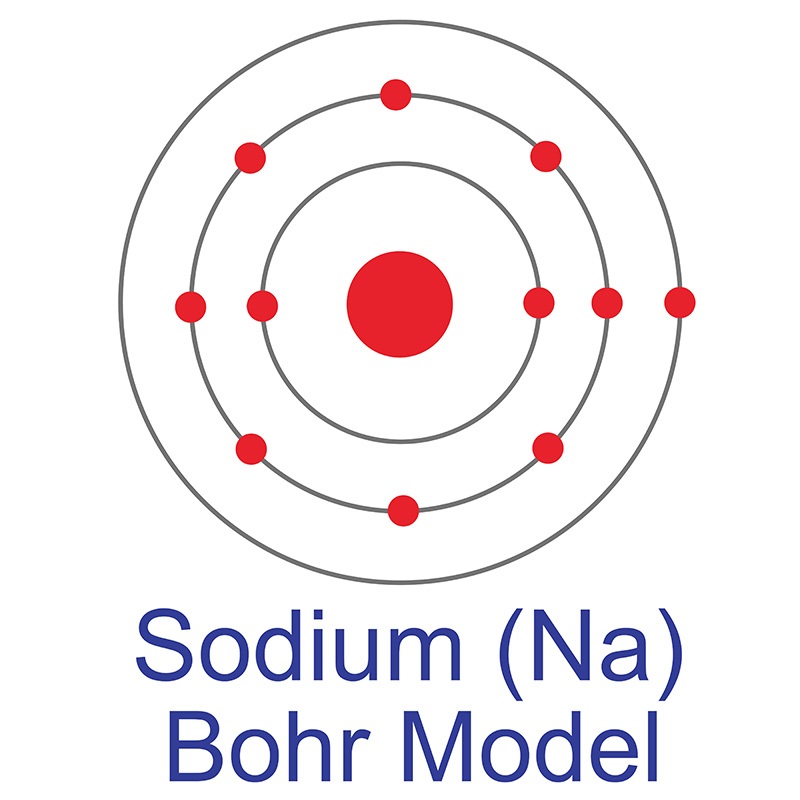
 See more Sodium products. Sodium (atomic symbol: Na, atomic number: 11) is a Block D, Group 5, Period 4 element with an atomic weight of 22.989769. The number of electrons in each of Sodium's shells is [2, 8, 1] and its electron configuration is [Ne] 3s1. The sodium atom has a radius of 185.8 pm and a Van der Waals radius of 227 pm. Sodium was discovered and first isolated by Sir Humphrey Davy in 1807. In its elemental form, sodium has a silvery-white metallic appearance. It is the sixth most abundant element, making up 2.6 % of the earth's crust. Sodium does not occur in nature as a free element and must be extracted from its compounds (e.g., feldspars, sodalite, and rock salt). The name Sodium is thought to come from the Arabic word suda, meaning "headache" (due to sodium carbonate's headache-alleviating properties), and its elemental symbol Na comes from natrium, its Latin name.
See more Sodium products. Sodium (atomic symbol: Na, atomic number: 11) is a Block D, Group 5, Period 4 element with an atomic weight of 22.989769. The number of electrons in each of Sodium's shells is [2, 8, 1] and its electron configuration is [Ne] 3s1. The sodium atom has a radius of 185.8 pm and a Van der Waals radius of 227 pm. Sodium was discovered and first isolated by Sir Humphrey Davy in 1807. In its elemental form, sodium has a silvery-white metallic appearance. It is the sixth most abundant element, making up 2.6 % of the earth's crust. Sodium does not occur in nature as a free element and must be extracted from its compounds (e.g., feldspars, sodalite, and rock salt). The name Sodium is thought to come from the Arabic word suda, meaning "headache" (due to sodium carbonate's headache-alleviating properties), and its elemental symbol Na comes from natrium, its Latin name.
Materials
Materials by Form
2D Materials Alloy & Alloy Forms Pure Metals & Metal FormsCeramic FibersFoams: Metallic & Ceramic High Purity Materials Isotopes MXenesOxides Rare Earths Semiconductors Solutions
Chemicals & Salts
All Chemicals & Salts Acetates Aluminides Ammonium Sulfates Antimonides Arsenates Benzoate Bromates Bromides Carbonates Chlorides Chromates Fluorides Hydrides Hydroxides Iodates Iodides Lactates Molybdates Nitrates Oxalates Oxides Perchlorates Phosphates Selenates Selenides Selenites Silicates Stearates Sulfates Sulfides Sulfites Tantalates Tellurates Tellurides Tellurites ThiocyanatesVanadates
Ceramics
Nanomaterials
Organometallics
Materials by Application
Additive Manufacturing & 3D Printing Battery & Supercapacitor Materials Catalysts Dental Materials Electronics Materials Fuel Cell Materials Fusion EnergyGlass Manufacturing Green Technology & Alternative Energy Hydrogen Storage Laser Crystals Life Sciences & Biomaterials Metallurgy Nanotechnology & Nanomaterials Optical Materials Photovoltaic & Solar Energy Plating Pigments & Coatings Research & Development Space Technology Sputtering Targets Thin Film Deposition Water Treatment Weather Modification
Life Science Chemicals
Life Science Products AlcoholsAldehydesAmidesAminesAmino Acids & DerivativesAromaticsArylsAzetidinesBenzimidazolesBenzisoxazolesBenzodioxansBenzofuransBenzothiazolesBenzothiophenesBenzoxazolesCarboxylic AcidsEnzymes & InhibitorsEstersEthersFluorinated Building BlocksFuransHalidesImidazolesImidazolidinesIndazolesIndolesIndolinesIsoquinolinesIsoxazolesKetonesMorpholinesNaphthyridinesNitrilesOrganoboronOrganosiliconOxadiazolesOxazolesPharmaceuticals & IntermediatesPhenolsPhytochemicalsPiperazinesPiperidinesPyrazinesPyrazolesPyridazinesPyridinesPyrimidinesPyrrolesPyrrolidinesPyrrolinesQuinazolinesQuinolinesQuinoxalinesSpiroesSulfonyl ChloridesTetrahydroisoquinolinesTetrahydropyransTetrahydroquinolinesTetrazolesThiadiazolesThiazolesThiazolidinesThiolsThiophenesTriazinesTriazoles
About Us
Locations
Austria Belgium Brazil Canada China & Hong Kong Czech Republic Denmark Finland France Germany Greece Hungary India Indonesia Israel Italy Japan Malaysia Mexico Netherlands Norway Philippines Poland Portugal Russia Singapore South Korea Spain Sweden Switzerland Taiwan Thailand Turkey United Kingdom United States
Industries
Aerospace Agriculture Automotive Chemical Manufacturing Defense Dentistry Electronics Energy Storage & Batteries Fine Art Materials Fuel CellsFusion Energy Glass Investment Grade Metals Jewelry & Fashion Lasers Lighting Medical Devices Museums & Galleries Nuclear Energy Oil & Gas Optics Paper & Pulp Pharmaceuticals & Cosmetics Research & Laboratory Robotics Solar Energy Space Sports Equipment Steel & Alloy Producers Textiles & Fabrics Water Treatment Municipalities
Follow Us
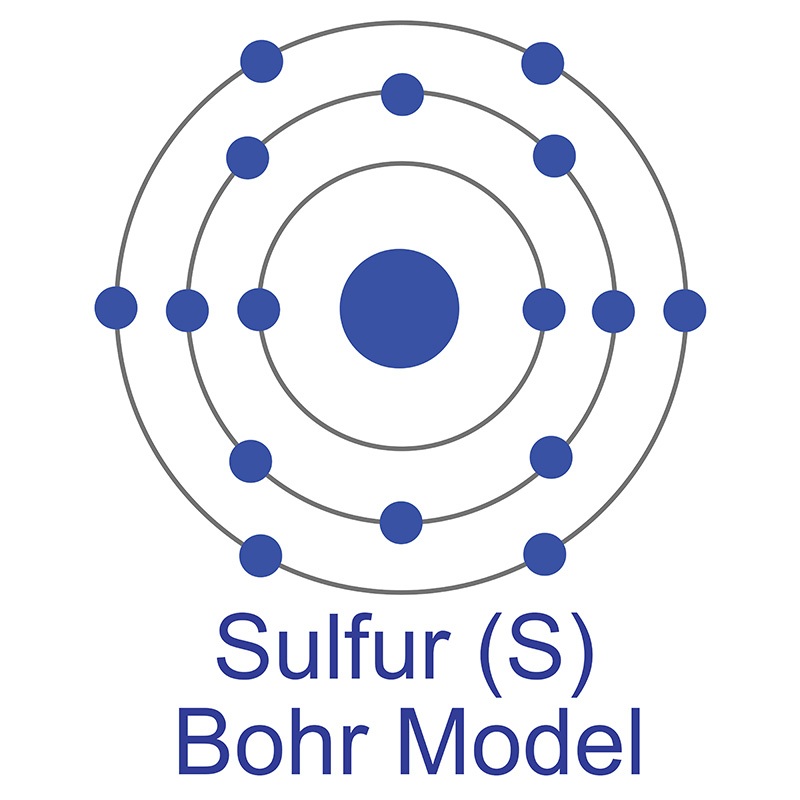 The number of electrons in each of Sulfur's shells is 2, 8, 6 and its electron configuration is [Ne] 3s2 3p4. In its elemental form, sulfur has a light yellow appearance. The sulfur atom has a covalent radius of 105 pm and a Van der Waals radius of 180 pm. In nature, sulfur can be found in hot springs, meteorites, volcanoes, and as galena, gypsum, and epsom salts. Sulfur has been known since ancient times but was not accepted as an element until 1777, when Antoine Lavoisier helped to convince the scientific community that it was an element and not a compound.
The number of electrons in each of Sulfur's shells is 2, 8, 6 and its electron configuration is [Ne] 3s2 3p4. In its elemental form, sulfur has a light yellow appearance. The sulfur atom has a covalent radius of 105 pm and a Van der Waals radius of 180 pm. In nature, sulfur can be found in hot springs, meteorites, volcanoes, and as galena, gypsum, and epsom salts. Sulfur has been known since ancient times but was not accepted as an element until 1777, when Antoine Lavoisier helped to convince the scientific community that it was an element and not a compound.